Structuring and storing data¶
The biological sciences are becoming more and more data driven. However, dealing with large volumes of data can feel overwhelming. One way to regain some control is to ensure that there is some structure to the data. This chapter introduces important data structures and file formats for representing them.
Data structures¶
In How to think like a computer we introduced some basic data types, including integers, floating point numbers, characters and strings. However, most of the time we are not interested in individual instances of integers or floating point values, we want to analyse lots of data points, often of a heterogeneous nature.
We need some way of structuring our data. In computing there are several well defined data structures for accomplishing this. Three data structures of particular interest are lists, dictionaries and graphs. Lists and dictionaries are generally useful and graphs are of particular interest in biology and chemistry.
A list, also known as an array, is basically a collection of elements in a specific order. The most common use case is to simply go through all the elements in a list and do something with them. For example if you had a stack of Petri dishes you might go through them, one-by-one, and only keep the ones that had single colonies suitable for picking. This is similar to what we did with the FASTA file in First steps towards automation, where we only retained the lines contain the expression “OS=Homo sapiens”.
Because lists have an order associated with them they can also be used to implement different types of queuing behaviour such as “first-in-first-out” and “first-in-last-out”. Furthermore, individual elements can usually be accessed through their numerical indices. This also means that you can sort lists. For example you could stack your Petri dishes based on the number of colonies in each one. Below is some pseudo code illustrating how a bunch of petri dish identifiers could be stored as a list.
petri_dishes = ["wt_control",
"wt_treated",
"mutant1_control",
"mutant1_treated",
"mutant2_control",
"mutant2_treated"]
In Python one can access the third element ("mutant1_control") using the syntax petri_dishes[2].
Note that index used is 2, rather than 3.
This is because Python uses zero-based indexing, i.e. the first element of
the list would be accessed using index zero, the third element therefore
has index two.
A dictionary, also known as a map or an associative array, is an unordered collection of elements that can be accessed through their associated identifiers. In other words each entry in a dictionary has a key, the identifier, and a value. For example, suppose that you needed to store information about the speed of various animals. In this case it would make a lot of sense to have a system where you could look up the speed information based on the animal’s name.
animal_speed = {"cheeta": 120,
"ostrich": 70,
"cat": 30,
"antelope": 88}
In Python one could look up the speed of an ostrich using the syntax
animal_speed["ostrich"].
It is worth noting that it is possible to create nested data structures. For example, think of a spreadsheet. The spreadsheet could be represented as a list containing lists. In other words the elements of the outer list would represent the rows in the spreadsheet and the elements in an inner list would represent values of cells at different columns for a specific row.
Let’s illustrate this using the table below.
| Time | Height | Weight |
|---|---|---|
| 0 | 0 | 0 |
| 5 | 2 | 5 |
| 10 | 7 | 12 |
| 15 | 8 | 20 |
The table, excluding the header, could be represented using a list of lists.
table = [[0, 0, 0],
[5, 2, 5],
[10, 7, 12],
[15, 8, 20]]
A graph, sometimes known as a tree, is a data structure that links nodes together via edges (Fig. 3). This should sound relatively familiar to you as it is the basic concept behind phylogenetic trees. Each node represents a species and the edges represent the inferred evolutionary relationships between the species. Graphs are also used to represent 2D connectivities of molecules.
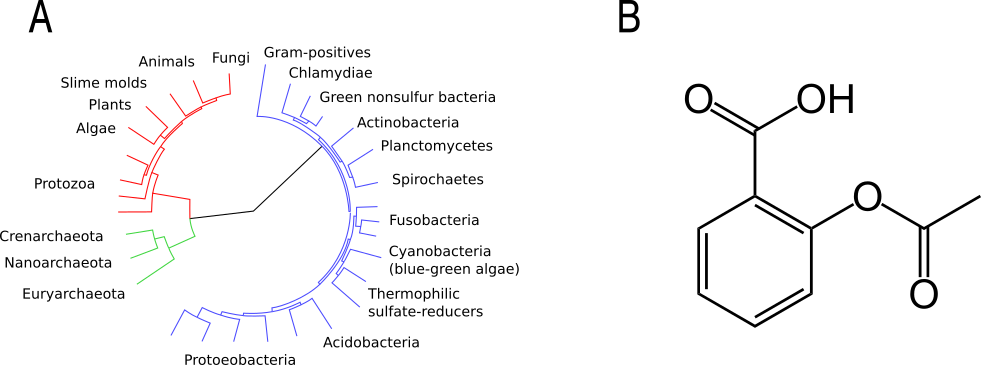
Fig. 3 Two examples of graphs: a phylogentic tree (A) and the chemical structure of Aspirin (B). Original images via Wikimeda Commons [Public domain] A and B.
Because of their general utility lists and dictionaries are built-in to many high level programming languages such as Python and JavaScript. However, data structures for graphs are generally not.
Data persistence¶
Suppose that your program has generated a phylogenetic tree and it has used this tree to determine that scientists and baboons are more closely related than expected. Result! At this point the program is about to finish. What should happen to the phylogenetic tree? Should it be discarded or should it be stored for future use? If you want to store data for future use you need to save the data to disk.
When you want to save data to disk you have a choice: you can save the data in a binary format that only your program understands or you can save the data as a plain text file. Storing your data in a binary format has advantages in that the resulting files will be smaller and they will load quicker than a plain text file. However, in the next section you will find out why you should (almost) always store your data as plain text files.
The beauty of plain text files¶
Plain text files have several advantages over binary format files. First of all you can open and edit them on any computer. The operating system does not matter as ASCII and Unicode are universal standards. With the slight caveat that you may occasionally have to deal with converting between Windows and Unix line endings (as discussed earlier in How to think like a computer).
Furthermore, they are easy to use. You can simply open them in your text editor of choice and start typing away.
Some software companies try to employ a lock-in strategy where their software produces files in a proprietary, binary format. Meaning that you need access to the software in order to open the files produced using it. This is not great from the users point of view. It makes it difficult to use other tools to further analyse the data. It also makes it hard to share data with people that do not have a licence to the software in question. Making use of plain text files and software that can output data in plain text works around this problem.
Finally, there is a rich ecosystem of tools available for working with plain
text files. Apart from text editors, there are all of the Unix command line
tools. We looked at some of these in First steps towards automation.
In the next chapter, Keeping track of your work, we will look at a
tool called git that can be used to track changes to plain text files.
Useful plain text file formats¶
There are many well established file formats for representing data in plain text. These have arisen to solve different types of problems.
Plain text files are commonly used to store notes, for example the minutes of a meeting or a list of ideas. When writing these types of documents one wants to be able to make use of headers, bullet points etc. A popular file format for creating such documents is markdown. Markdown (MD) provides a simple way to add formatting such as headers and bullet lists by providing a set of rules of for how certain plain text constructs should be converted to HTML and other document formats.
# Level 1 header
## Level 2 header
Here is some text in a paragraph.
It is possible to *emphasize words with italic*.
It is also possible to **strongly emphansize words in bold**.
- First item in a bullet list
- Second item in a bullet list
1. First item in a numbered list
2. Second item in a numbered list
[Link to BBC website](www.bbc.com)

Hopefully the example above is self explanatory. For more information have a look at the official markdown syntax page.
Another scenario is to record tabular data, for example the results of a scientific experiment. In other words the type of data you would want to store in a spreadsheet. Comma Separated Value (CSV) files are ideally suited for this. This file format is relatively basic, values are simply separated by commas and the file can optionally start with a header. It is worth noting that you can include a comma in a value by surrounding it by double quotes. Below is an example of a three column CSV file containing a header and two data rows.
Last name,First name(s),Age
Smith,Alice,34
Smith,"Bob, Carter",56
Another scenario, when coding, is the ability to store richer data structures, such as lists or dictionaries, possibly nested within each other. There are two popular file formats for doing this JavaScript Object Notation (JSON) and YAML Ain’t Markup a Language (YAML).
JSON was designed to be easy for machines to generate and parse and is used extensively in web applications as it can be directly converted to JavaScript objects. Below is an example of JSON representing a list of scientific discoveries, where each discovery contains a set of key value pairs.
[
{
"year": 1653,
"scientist": "Robert Hooke",
"experiment": "light microscopy",
"discovery": "cells"
},
{
"year": 1944,
"scientist": "Barbara McClintock",
"experiment": "breeding maize plants for colour",
"discovery": "jumping genes"
}
]
YAML is similar to JSON in that it is a data serialisation standard. However, it places more focus on being human readable. Below is the same data structure represented using YAML.
---
-
year: 1653
scientist: "Robert Hooke"
experiment: "light microscopy"
discovery: "cells"
-
year: 1944
scientist: "Barbara McClintock"
experiment: "breeding maize plants for colour"
discovery: "jumping genes"
A nice feature of YAML is the ability to add comments to the data giving further explanation to the reader. These comments are ignored by programs parsing the files.
---
# TODO: include an entry for Anton van Leeuwenhoek here.
-
year: 1653
scientist: "Robert Hooke"
experiment: "light microscopy"
discovery: "cells"
-
year: 1944
scientist: "Barbara McClintock"
experiment: "breeding maize plants for colour"
discovery: "jumping genes"
As scientist’s we sometimes need to be able to work with graph data, for example phylogenetic trees and molecules. These often have their own domain specific plain text file formats. For example the Newick format is commonly used to store phylogenetic trees and there are a multitude of file formats for representing molecules including the SMILES, and Molfile file formats.
A neat file format for storing and visualising generic graph data is the DOT language. Plain text files written in the DOT language can be visualised using the software Graphviz.
Some figures are well suited for being stored as plain text files. This is the case when all the content of the graphic can be described as mathematical functions. These are so called vector graphics and the standard file format for storing them as plain text files is Scalable Vector Graphics. A neat feature of these types of images is that they can be arbitrarily scaled to different sizes without losing any resolution. Hence the word “scalable” in their name.
However, there is another type of graphic that is ill suited to being represented as plain text files. These are so called raster images. In raster images the graphic is represented as a grid where each grid point is set to a certain intensity. Common file formats include PNG, JPEG, and GIF. If you are dealing with photographs or microscopy images the raw data will be recorded in raster form. The reason for storing these types of images as binary blobs, rather than plain text files, is that it saves a significant amount of disk space. Furthermore, image viewers can load these binary blobs much quicker than they could load the same amount of information stored as a plain text file.
However, suppose you needed to generate a figure as a raster image, say for example a scatter plot. Then you should consider writing a script to generate the figure. The instructions for generating the figure, i.e. the script, can then be stored as a plain text file. This concept will be explored in Data visualisation.
Tidy data¶
In the Data visualisation chapter we will make use of the ggplot2 package. This requires data to be structured as Tidy Data, where each variable is a column and each observation is a row and each type of observational unit forms a table.
Take for example the table below.
| Control | Heat shock | |
|---|---|---|
| Wild type | 3 | 15 |
| Mutant | 5 | 16 |
This data would be classified as “messy” because each row contains two observations, i.e. the control and the heat shock experiments. To reorganise the data so that it becomes tidy we need to “melt” or stack it.
| Variant | Experiment | Result |
|---|---|---|
| Wild type | Control | 3 |
| Wild type | Heat shock | 15 |
| Mutant | Control | 5 |
| Mutant | Heat shock | 16 |
The benefit of structuring your data in a tidy fashion is that it makes it easier to work with when you want to visualise and analyse it.
Find a good text editor and learn how to use it¶
A key step to boost your productivity is to find a text editor that suits you, and learning how to make the most of it.
Popular text editors include Sublime Text, Geany and Atom. I would recommend trying out at least two of them and doing some of your own research into text editors. Editing text files will be a core activity throughout the rest of this book and you want to be working with a text editor that makes you happy!
If you enjoy working on the command line I would highly recommend experimenting with command line text editors. Popular choices include nano, emacs and vim. The former is easy to learn, whereas the latter two give much more power, but are more difficult to learn.
Key concepts¶
- Lists, also known as arrays, are ordered collections of elements
- Dictionaries, also known as maps and associative arrays, are unordered collections of key-value pairs
- Graphs, sometimes known as trees, links nodes via edges and are of relevance to phylogenetic trees and molecular representations
- In computing persistence refers to data outliving the program that generated it
- If you want any data structures that you have generated to persist you need to write them to disk
- Saving your data as plain text files is almost always preferable to saving it as a binary blob
- There are a number of useful plain text file formats for you to make use of
- Don’t invent your own file format
- Structuring your data in a “tidy” fashion will make it easier to analyse and visualise
- Learn how to make the most out of your text editor of choice